Image Synthesis in Neuroimaging
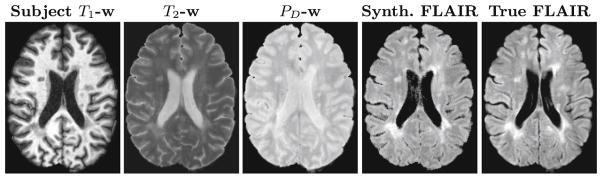 FLAIR Image Synthesis
FLAIR Image SynthesisAutomatic processing of magnetic resonance images (MRI) is a vital part of neuroscience research. Yet even the best and most widely used medical image processing methods will not produce consistent results when their input images are acquired with different pulse sequences. The lack of consistency is a result of multiple sources of variation in the acquired MRI data. MRI, unlike computed tomography (CT), does not produce images where the magnitude of the intensity is standardized across scanners. In a typical scanning session, different MRI pulse sequences are acquired at different resolutions for various reasons. Certain pulse sequences are prone to artifacts that cause corrupted data. Image synthesis has been proposed to enhance and/or homogenize acquired MRI data. We have developed three different approaches to perform image synthesis in MRI:
- PSICLONE
- REPLICA
- SynthCRAFT
PSICLONE
PSICLONE (Pulse Sequence Information-based Contrast Learning On Neighborhood Ensembles) is an image synthesis and intensity standardization framework.
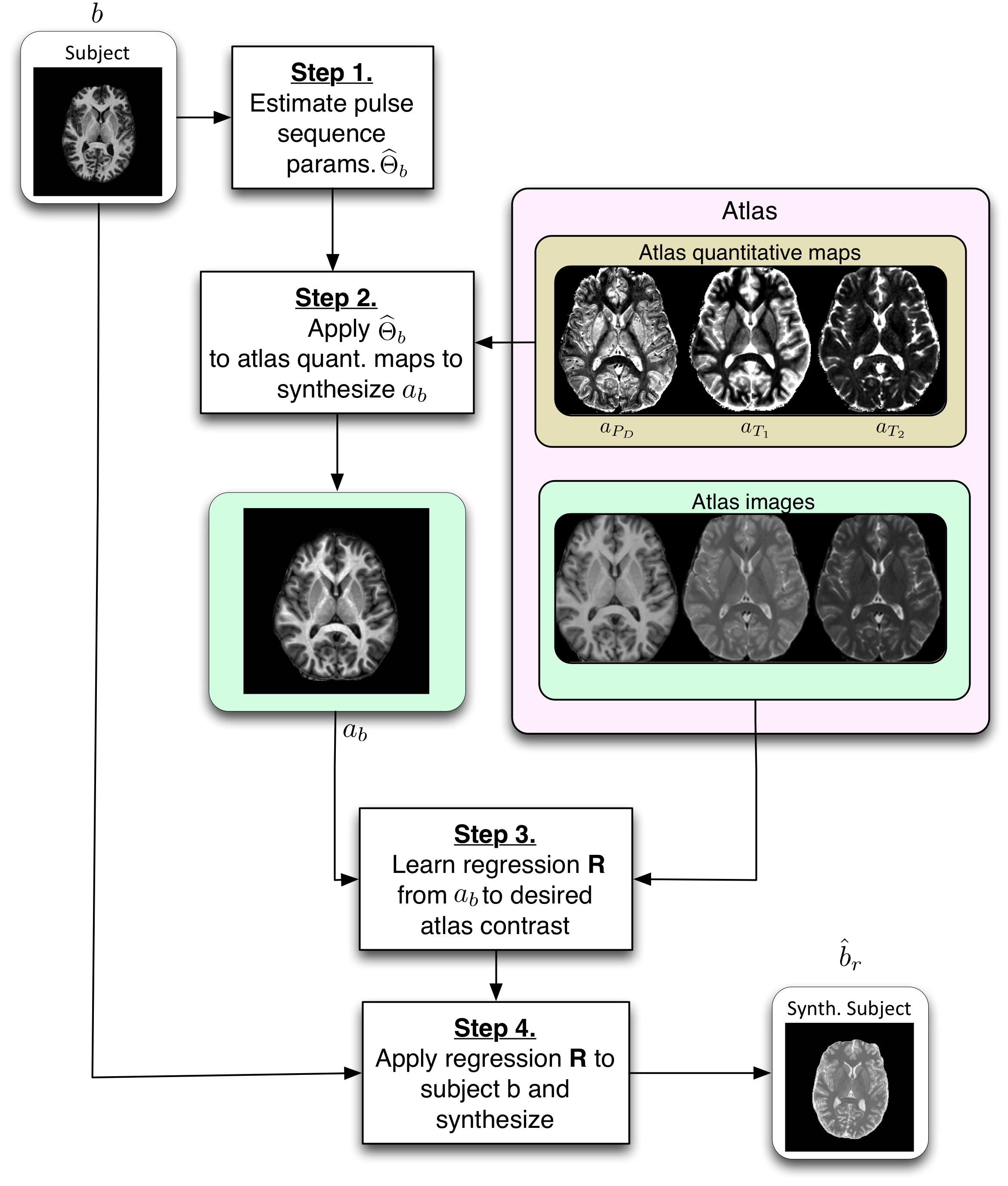
PSICLONE estimates approximate pulse sequence parameters of a given MR image to generate subject-specific training images that are used to train a random forest regression to synthesize a target contrast. The learnt regression is applied to the given image to generate a synthetic or a standardized image.
Paper: doi
REPLICA
REPLICA (Regression Ensembles with Patch Learning for Image Contrast Agreement) is a supervised, multi-resolution image synthesis approach that uses random forests as the main regression engine. REPLICA can be used as an image synthesis approach on its own as well as a subroutine in PSICLONE.
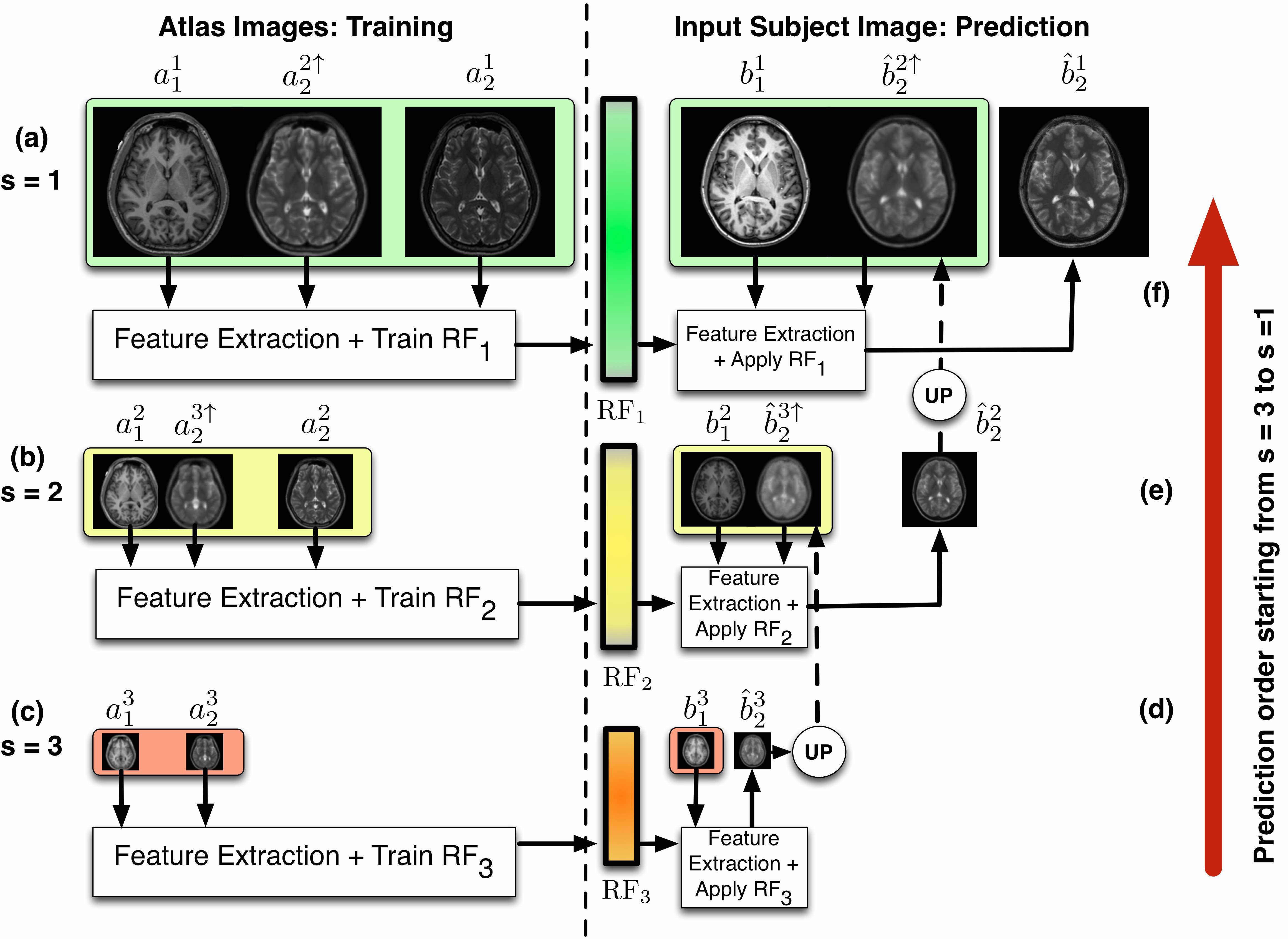
Paper: REPLICA: Regression Ensembles with Patch Learning for Image Contrast Agreement
Code: REPLICA MATLAB code
SynthCRAFT
In SynthCRAFT (Synthesis with Conditional Random Field Tree), we frame the image synthesis problem as an inference problem on a 3-D continuous-valued conditional random field (CRF). We model the conditional distribution as a Gaussian by defining quadratic association and interaction potentials encoded in leaves of a regression tree. The parameters of these quadratic potentials are learned by maximizing the pseudo-likelihood of the training data. Final synthesis is done by inference on this model.
Paper: IPMI paper